Example: Autocorrelator of a Hermitian operator¶
Observable kind: BosonAutoCorr.
Correlator of a Hermitian operator with itself.
For the Hermitian operators the auxiliary spectral function \(A(\epsilon) = \Im\chi(\epsilon)/\epsilon\) is non-negative and symmetric.
This mode is faster and more robust than BosonCorr.
Run analytical continuation¶
# Import some TRIQS modules and NumPy
from pytriqs.gf.local import *
from pytriqs.archive import HDFArchive
import pytriqs.utility.mpi as mpi
import numpy
# Import main SOM class
from pytriqs.applications.analytical_continuation.som import Som
n_w = 501 # Number of energy slices for the solution
energy_window = (0,2.5) # Energy window to search the solution in
# The window must entirely lie on the positive half-axis
# Parameters for Som.run()
run_params = {'energy_window' : energy_window}
# Verbosity level
run_params['verbosity'] = 3
# Number of particular solutions to accumulate
run_params['l'] = 5000
# Number of global updates
run_params['f'] = 100
# Number of local updates per global update
run_params['t'] = 50
# Accumulate histogram of the objective function values
run_params['make_histograms'] = True
# Read \chi(i\omega_n) from archive
# Could be \chi(\tau) or \chi_l as well.
chi_iw = HDFArchive('example.h5', 'r')['chi_iw']
# Set the weight function S to a constant (all points of chi_iw are equally important)
S = chi_iw.copy()
S.data[:] = 1.0
# Estimated norms of spectral functions, (\pi/2) * \chi(i\omega_0)
norms = (numpy.pi/2) * numpy.array([chi_iw(0).real[0,0],
chi_iw(0).real[1,1]])
# Construct a SOM object
cont = Som(chi_iw, S, kind = "BosonAutoCorr", norms = norms)
# Run!
# Takes 2-3 minutes on 4 cores ...
cont.run(**run_params)
# Evaluate the solution on an energy mesh
# NB: we can use *any* energy window at this point, not necessarily that from run_params
chi_w = GfReFreq(window = (0,2.5), n_points = n_w, indices = [0,1])
chi_w << cont
# \chi(i\omega_n) reconstructed from the solution
chi_rec_iw = chi_iw.copy()
chi_rec_iw << cont
# On master node, save results to an archive
if mpi.is_master_node():
with HDFArchive("results.h5",'w') as ar:
ar['chi_iw'] = chi_iw
ar['chi_rec_iw'] = chi_rec_iw
ar['chi_w'] = chi_w
ar['histograms'] = cont.histograms
Download input file example.h5.
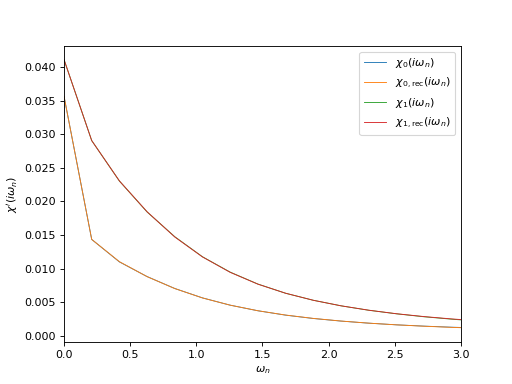
Plot input and reconstructed correlators at Matsubara frequencies¶
from pytriqs.gf.local import *
from pytriqs.archive import HDFArchive
from matplotlib import pyplot as plt
from pytriqs.plot.mpl_interface import oplot
# Read data from archive
ar = HDFArchive('results.h5', 'r')
# Plot input and reconstructed \chi(i\omega_n)
oplot(ar['chi_iw'][0,0], mode='R', linewidth=0.8, label="$\chi_0(i\\omega_n)$")
oplot(ar['chi_rec_iw'][0,0], mode='R', linewidth=0.8, label="$\chi_\mathrm{0,rec}(i\\omega_n)$")
oplot(ar['chi_iw'][1,1], mode='R', linewidth=0.8, label="$\chi_1(i\\omega_n)$")
oplot(ar['chi_rec_iw'][1,1], mode='R', linewidth=0.8, label="$\chi_\mathrm{1,rec}(i\\omega_n)$")
plt.xlim((0, 3))
plt.ylabel("$\chi'(i\\omega_n)$")
plt.legend(loc="upper right")
(Source code, png, hires.png, pdf)
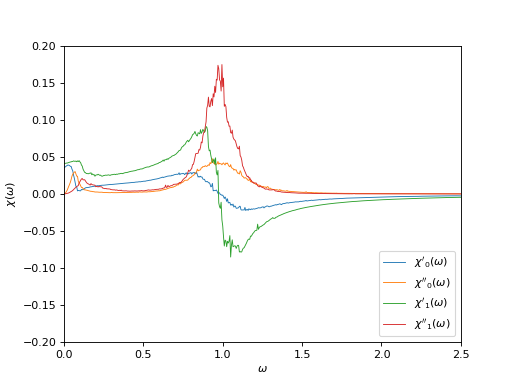
Plot the correlator on the real frequency axis¶
from pytriqs.gf.local import *
from pytriqs.archive import HDFArchive
from matplotlib import pyplot as plt
from pytriqs.plot.mpl_interface import oplot
# Read data from archive
ar = HDFArchive('results.h5', 'r')
# Plot the susceptibility on the real axis
oplot(ar['chi_w'][0,0], mode='R', linewidth=0.8, label="$\chi'_0(\\omega)$")
oplot(ar['chi_w'][0,0], mode='I', linewidth=0.8, label="$\chi''_0(\\omega)$")
oplot(ar['chi_w'][1,1], mode='R', linewidth=0.8, label="$\chi'_1(\\omega)$")
oplot(ar['chi_w'][1,1], mode='I', linewidth=0.8, label="$\chi''_1(\\omega)$")
plt.xlim((0,2.5))
plt.ylim((-0.2,0.2))
plt.ylabel("$\chi(\\omega)$")
plt.legend(loc = "lower right")
(Source code, png, hires.png, pdf)